general-purpose approaches to learn functional relationships from data without the need to define them a priori derive predictive models without the need for strong assumptions about underlying mechanisms, which are frequently unknown or insufficiently defined (especially for genomic data) he most accurate prediction of gene expression levels is currently made from a broad set of epigenetic features using linear models or random forests However, how the selected features determine the transcript levels remains an active research topic Most of machine learning applications in genomics can be described within the canonical machine learning workflow with four steps data cleaning and pre-processing feature extraction model fitting/training evaluation It is customary to denote one data sample, including all covariates and features as input x (usually a vector of numbers), and label it with its response variable or output value y (usually a single number) when available. approaches aim to discover patterns from the data samples x themselves, without the need for output labels y. Methods such as clustering and principal component analysis are typical examples of unsupervised models applied to biological data 深度神经网络 An artificial neural network, initially inspired by neural networks in the brain, consists of layers of interconnected compute units (neurons). The depth of a neural network corresponds to the number of hidden layers, and the width to the maximum number of neurons in one of itslayers. When training networks with larger numbers of hidden layers, artificial neural networks were rebranded as “deep networks”. The term “neural network” largely refers to the hypothesis class part of a machine learning algorithm: architectures Recurrent neural networks application CNN architectures to predict specificities of DNA-binding and RNAbinding proteins Outperformed existing methods, to recover known and novel sequence motifs, and could quantify the effect of sequence alterations and identify functional SNVs The neurons in the convolutional layer scan for motif sequences and combinations thereof, similar to conventional PWMs The learning signal from deeper layers informs the convolutional layer which motifs are the most relevant. The motifs recovered by the model can then be visualized as sequence logos paper Need sufficient amount of labelled data Need to be trained, selected and tested on independent data sets to avoid overfitting and assure that the model will generalize The training set is used to learn models with different parameters, which are then assessed on the validation set. The model with best performance, for example prediction accuracy or meansquared error, is selected and further evaluated on the test set to quantify the performance on unseen data and for comparison to other methods. Typical data set proportions are 60% for training, 10% for validation and 30% for model testing. If the data set is small, k-fold cross-validation can be used Categorical features such as DNA nucleotides need to be encoded numerically. Typically represented as binary vectors with all but one entry set to zero (one-hot coding). The four bits of each encoded base are commonly considered analogously to color channels of an image to preserve the entity of a nucleotide 本文作者:思考问题的熊 版权声明:本博客所有文章除特别声明外,均采用 知识共享署名-非商业性使用-禁止演绎 4.0 国际许可协议 (CC BY-NC-ND 4.0) 进行许可。 如果你对这篇文章感兴趣,欢迎通过邮箱订阅我的 「熊言熊语」会员通讯,我将第一时间与你分享肿瘤生物医药领域最新行业研究进展和我的所思所学所想,点此链接即可进行免费订阅。recap
Machine learning methods
Advantage
Example
four steps
Supervised machine learning model
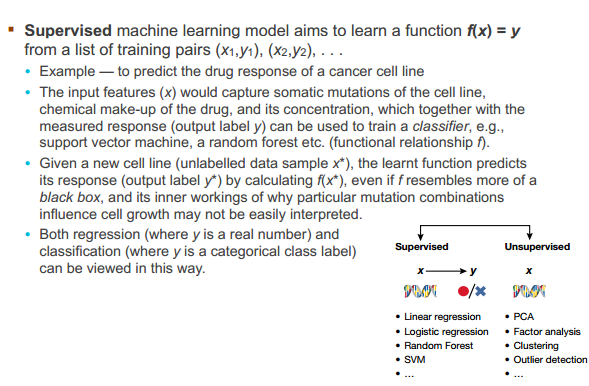
Unsupervised machine learning
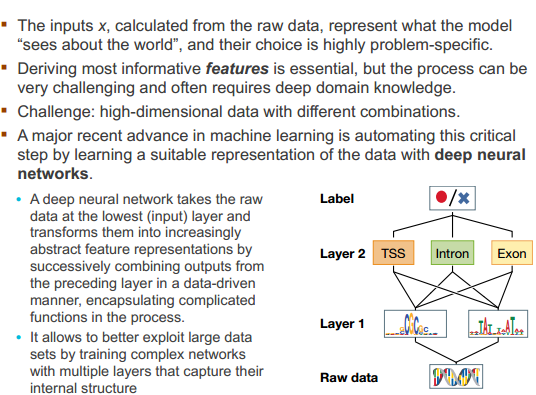
Neural networks

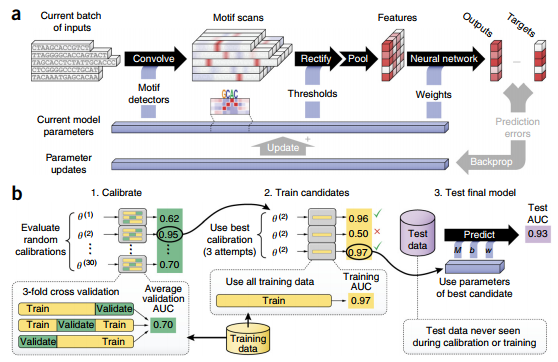
DeepBind
Important considerations related to data
· 分享链接 https://kaopubear.top/blog/2017-08-15-longxing-bioinfo-mlinbioinfo/