DOI(url): https://doi.org/10.1016/j.cell.2019.03.008 发表日期:4 April 2019 反义 lncRNA 转录可以影响 DNA 甲基化,进而改变染色体结构促进增强子与启动子结合,调控基因表达。 Pcdhα 基因有 13 个可以随机启动的可变外显子,每个启动子都可以由自身的启动子驱动,还有 3 个 c 型外显子以及稳定表达的编码 pcdh 结构域的外显子,同时,还有一个 enhancer 调控。 研究者发现,反义 lncRNA 的转录,会造成该位点 DNA 去甲基化的发生,从而使远端增强子靠近该外显子的启动子,促进它的表达。如图 2 所示,Pcdhα 基因座位均带有抑制表达的 DNA 甲基化修饰,而当反义 lncRNA 表达时,该外显子附近的 DNA 被 DNA 去甲基化酶 TET3 识别,去除了甲基化修饰,cohesin 蛋白重塑了染色体的结构,HS5-1 增强子与该基因座位的启动子结合,启动相应 Pcdhα 变体的表达。 关于反义 lncRNA 影响正义链基因表达的作用机制,主要有 3 类 CTCF: CTCF is an enhancer-blocking protein that inhibits the access of Igf2 to the enhancer elements located downstream from the H19 transcription start site. Cohesin Cohesin is a multiprotein complex that holds sister chromatids together from S phase until the start of mitosis, helping to ensure genomic integrity (Haarhuis, Elbatsh, & Rowland, 2014). 另一篇植物中相关 lncRNA 鉴定方法 First, only transcripts with TAIR10 annotation [Cufflinks class codes ‘u’ (intergenic transcripts),’x’ (Exonic overlap with reference on the opposite strand),’i’ (transcripts entirely within intron) were retained. Second, transcripts of short length (length <150 nt) or low abundance (FPKMmax < 1, FPKMmax stands for the maximum expression level of a lncRNA from all samples) were removed. Third, transcripts with protein-coding potential were removed. Protein-coding potential was determined by using two programs: (1) transcripts were subjected to a BlastX search against all plant protein sequences in the Swiss-Prot database70 with a cutoff e-value < 10-4 and the transcripts with strong hits (alignment length ≥40 aa, percent identity ≥35% and coverage of the alignment region in either query or subject sequence ≥35%) to known proteins were considered to have protein-coding potential; For antisense transcripts, open reading frames were checked. (2) the CPC (Coding Potential Calculator) score71, a value to assess protein-coding potential of a transcript based on six biologically meaningful sequence features, was calculated for each transcript. When the CPC score is positive, we considered the transcript to have protein-coding potential. Transcripts that passed the three filtering steps were annotated as lncRNAs. DOI(url): https://doi.org/10.1016/j.cels.2019.03.004 发表日期:April 4, 2019 一套分析 exRNA 的完整方案 exRNA 就是细胞外 RNA,也就是在细胞内转录但是在细胞外发挥功能?最近 cell 有一个专刊介绍了很多 exRNA 的文章,我扫了一眼,感觉主要内容是小 RNA 居多。所以这个分类和 lncRNA 类似,一个从长度一个从位置来进行区分。既然是这样,exRNA 的处理方法应该就和小 RNA 以及一般的 RNAseq 分析类似。看看这个流程里面有哪些不一样的地方。这里提供的分析方法从内容来看主要针对小 RNA,但是官方说也可以很方便的移植到其它 RNA,在 GitHub 的代码里也有针对 longRNA 的脚本。主要流程如下图,质控后会同时比对到多个数据库,具体内容可以参考 GitHub。 什么是 exRNA exRNA: Extracellular RNA (also known as exRNA or exosomal RNA) describes RNA species present outside of the cells from which they were transcribed. In Homo sapiens, exRNAs have been discovered in bodily fluids such as venous blood, saliva, breast milk, urine, semen, menstrual blood, and vaginal fluid. Although their biological function is not fully understood, exRNAs have been proposed to play a role in a variety of biological processes including syntrophy, intercellular communication, and cell regulation. exRNA 的种类 Extracellular RNA should not be viewed as a category describing a set of RNAs with a specific biological function or belonging to a particular RNA family. Similar to the term "non-coding RNA", "extracellular RNA" defines a group of several types of RNAs whose functions are diverse, yet they share a common attribute which, in the case of exRNAs, is existence in an extracellular environment. The following types of RNA have been found outside the cell: 研究 exRNA 的关键似乎应该是如何分离得到确实是细胞外的 RNA。 比如 Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell. 2019 Apr 4;177(2):446-462.e16. doi: 10.1016/j.cell.2019.03.024. 这篇文章就比较了集中 exRNA Isolation Methods,通过对 5 种生物液体中 10 种 exRNA 分离方法的系统比较,发现所得到的小 RNA-seq 图谱的复杂性和重现性存在显著差异。每种方法对不同的 exRNA 载体亚类的相对效率是通过估计细胞外囊泡 (EV)-、核糖核蛋白(RNP)- 和高密度脂蛋白(HDL) 特异性 miRNA 在每个图谱中的比例来确定的。开发了一种基于 web 的交互式应用(miRDaR),帮助研究人员为他们的研究选择最佳的 exRNA 分离方法。 另外,还有一篇文章:exRNA Atlas Analysis Reveals Distinct Extracellular RNACargo Types and Their Carriers Present across Human Biofluids. Cell. 2019 Apr 4;177(2):463-477.e15. doi: 10.1016/j.cell.2019.02.018. exRNA Atlas resource 包含来自 19 项研究的 5309 个 exRNA-seq 和 exRNAqPCR 概要文件,以及一套分析和可视化工具。通过分析,该研究得到了一个包含六种 exRNA 类型 (CT1、CT2、CT3A、CT3B、CT3C、CT4) 的模型,每种 exRNA 类型都可以在多种生物体液 (血清、血浆、脑脊液、唾液、尿液) 中检测到。 DOI(url): https://doi.org/10.1186/s12859-019-2745-1 发表日期:29 March 2019 不同物种之间 RNA-seq 怎么分析确实是一个问题,那么 ChIP-seq 呢? propose a scale based normalization (SCBN) method by taking into account the available knowledge of conserved orthologous genes and by using the hypothesis testing framework. Considering the different gene lengths and unmapped genes between different species, we formulate the problem from the perspective of hypothesis testing and search for the optimal scaling factor that minimizes the deviation between the empirical and nominal type I errors. 用小鼠来研究人的疾病非常常见,在一些文章中也有人会用同源基因进行比较。对于不同物种的数据来说,除了基因数量基因长度的不同,也有测序深度的问题。让两个物种之间的数据可比,数据的标准化方法非常重要。在之前的一些研究中,有人使用 RPKM 值来进行比较找到一千个保守基因,然后评估每个基因在不同物种中的中位数水平,然后通过让中位值保持一致来得到一个校正因子(median method)。这篇文章作者利用直系同源基因通过对已有方法的改进来进行不同物种之间数据的矫正。 ChIP-seq 类的数据有哪些方法呢? 雅卡尔指数(英语:Jaccard index),又称为并交比(Intersection over Union)、雅卡尔相似系数(Jaccard similarity coefficient),是用于比较样本集的相似性与多样性的统计量。雅卡尔系数能够量度有限样本集合的相似度,其定义为两个集合交集大小与并集大小之间的比例。在 bedtools 中有这个工具可以 对 jaccard 进行计算。 还有 余弦相似度 cosine similarity score ,比如这篇文章 A Cosine Similarity-Based Method to Infer Variability of Chromatin Accessibility at the Single-Cell Level 另外,dpca 可以用于分析转录因子结合位点处和启动子的不同染色质模式,以及等位基因特异性蛋白 - DNA 之间的相互作用。 DOI(url): https://doi.org/10.1093/bib/bbz039 发表日期:09 April 2019 难得的植物 DNA 甲基化分析综述文章 在这篇综述中,作者概述了分析 DNA 甲基化数据(特别是亚硫酸氢盐测序数据)最常用的生物信息学工具,也分析了这些工具的性能并且比较了计算拟南芥以及小麦甲基数据的计算时间和一致性。同时举例说明了作物中 DNA 甲基化数据分析的应用。但从软件上看,BSMap 用是最短,尤其是当线程数上去之后,但是内存则是 Bismark 最省。 关于内存的使用情况,不要被下图迷惑。小麦那里只是展示了处理 1 条染色体的需要的内存用量。要知道,小麦可是有 21 条染色体,16G 的基因组。 另外一篇文章,Strategies for analyzing bisulfite sequencing data 。 本文作者:思考问题的熊 版权声明:本博客所有文章除特别声明外,均采用 知识共享署名 - 非商业性使用 - 禁止演绎 4.0 国际许可协议 (CC BY-NC-ND 4.0) 进行许可。 如果你对这篇文章感兴趣,欢迎通过邮箱订阅我的 「熊言熊语」会员通讯,我将第一时间与你分享肿瘤生物医药领域最新行业研究进展和我的所思所学所想,点此链接即可进行免费订阅。Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice
关键点
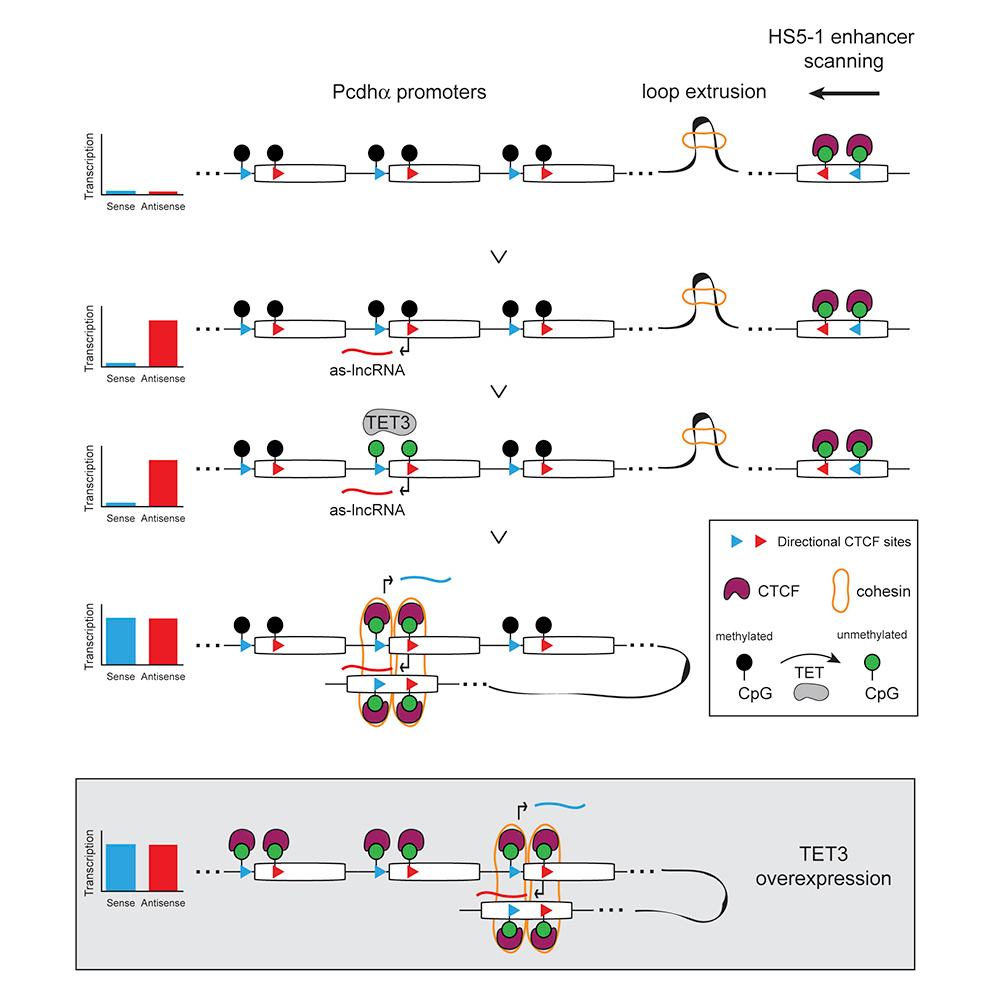
参考意义
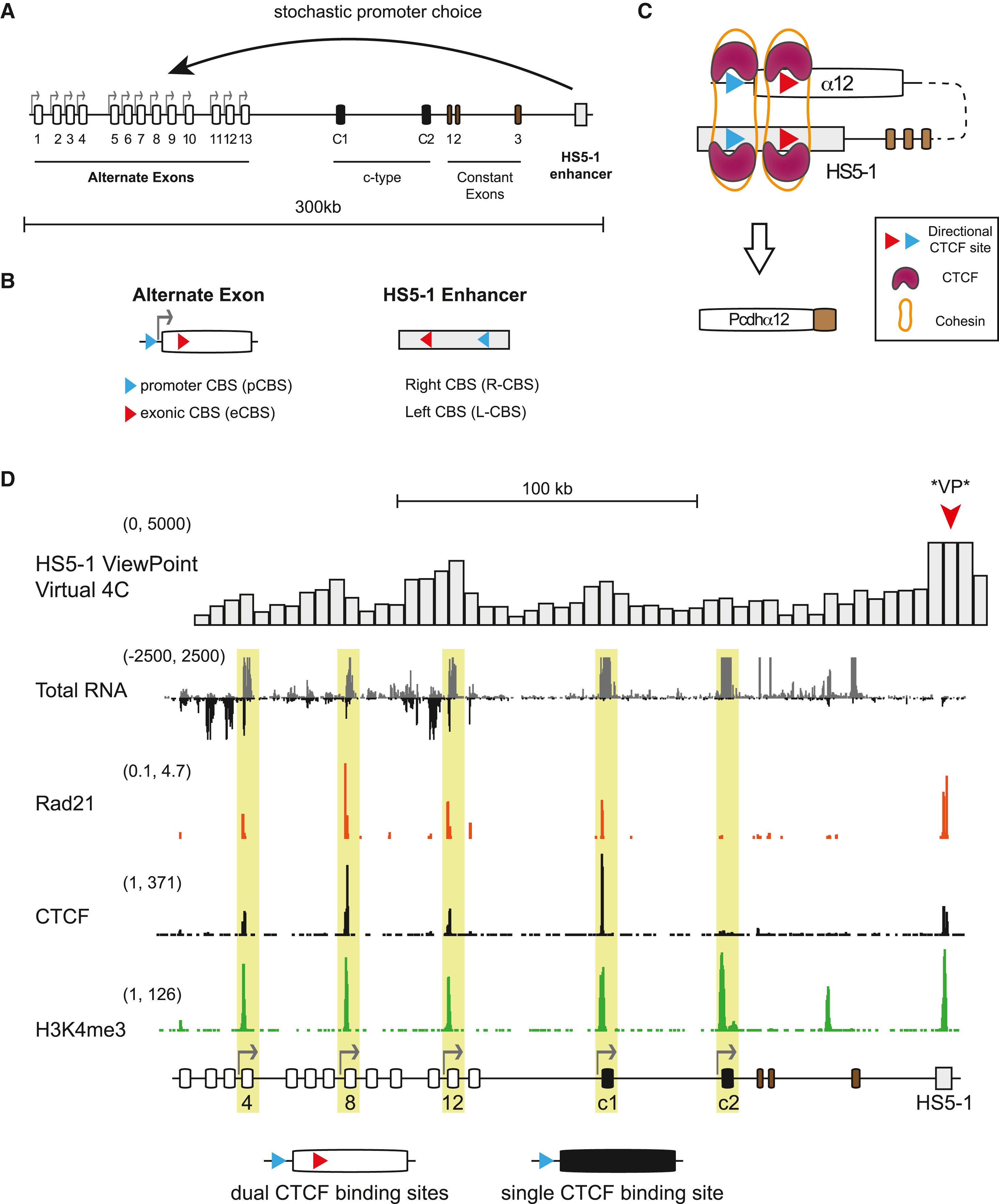
相关内容
exceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling
关键点
参考意义
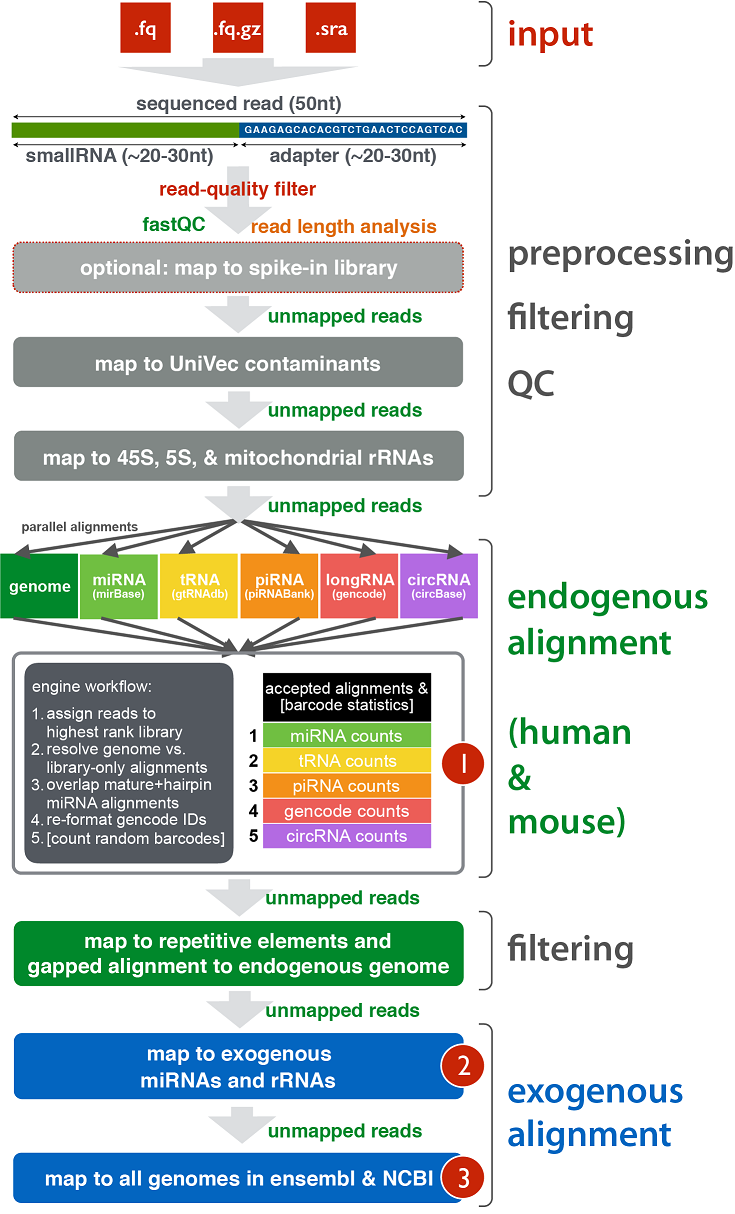
相关内容
A statistical normalization method and differential expression analysis for RNA-seq data between different species
关键点
参考意义
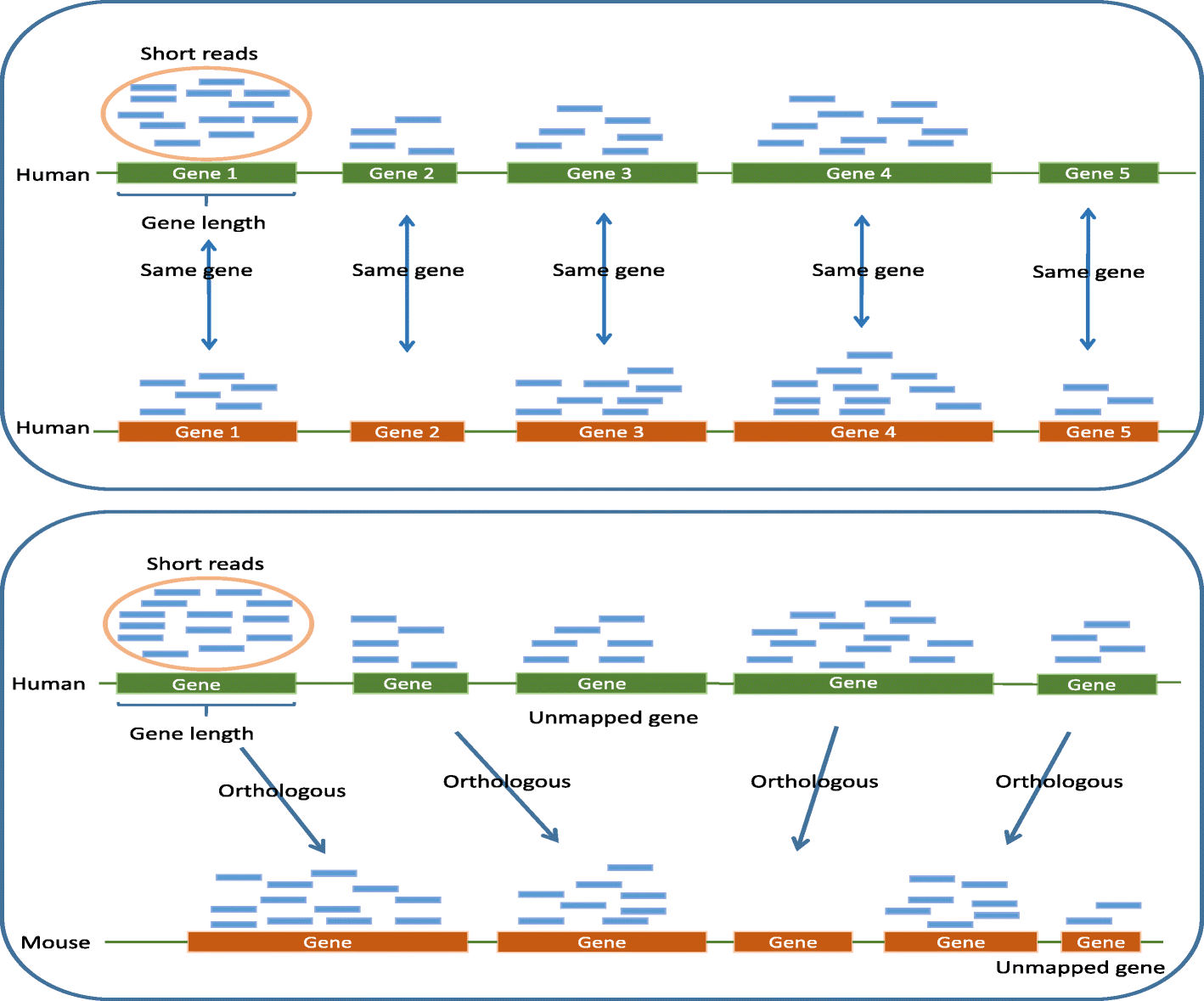
相关内容
DNA methylation analysis in plants: review of computational tools and future perspectives
关键点
参考意义
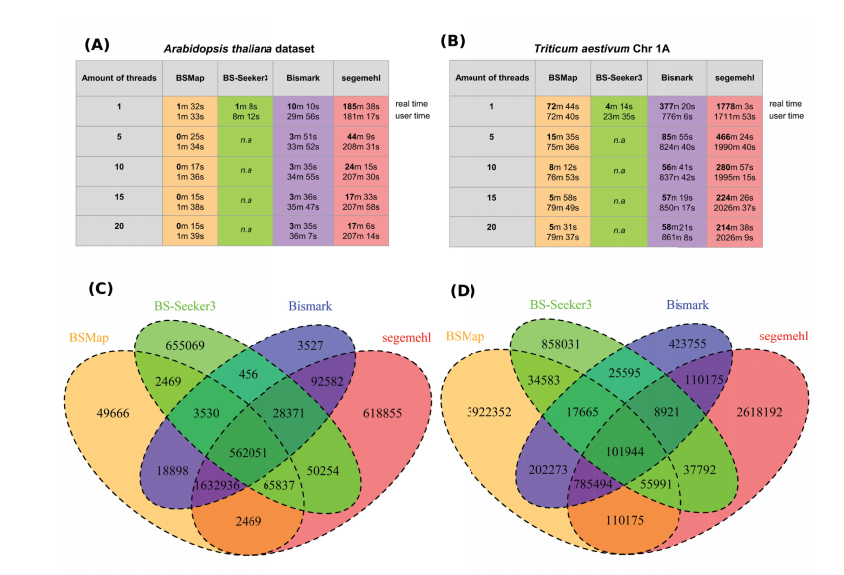
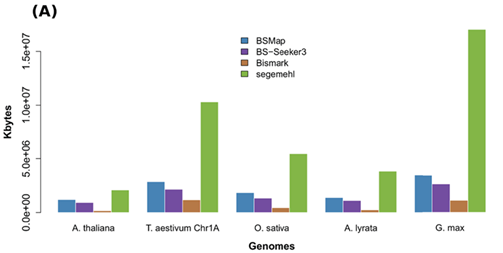
相关内容
· 分享链接 https://kaopubear.top/blog/2019-04-11-weeklypaper/